How to submit Spark jobs to EMR cluster from Airflow
Table of Contents
Introduction
I have been asked and seen the questions
how others are automating apache spark jobs on EMR
how to submit spark jobs to an EMR cluster from Airflow ?
In this post we go over the Apache Airflow way to
- Create an AWS EMR cluster.
- Submit Apache Spark jobs to the cluster using EMR’s Step function from Airflow.
- Wait for completion of the jobs.
- Terminate the AWS EMR cluster.
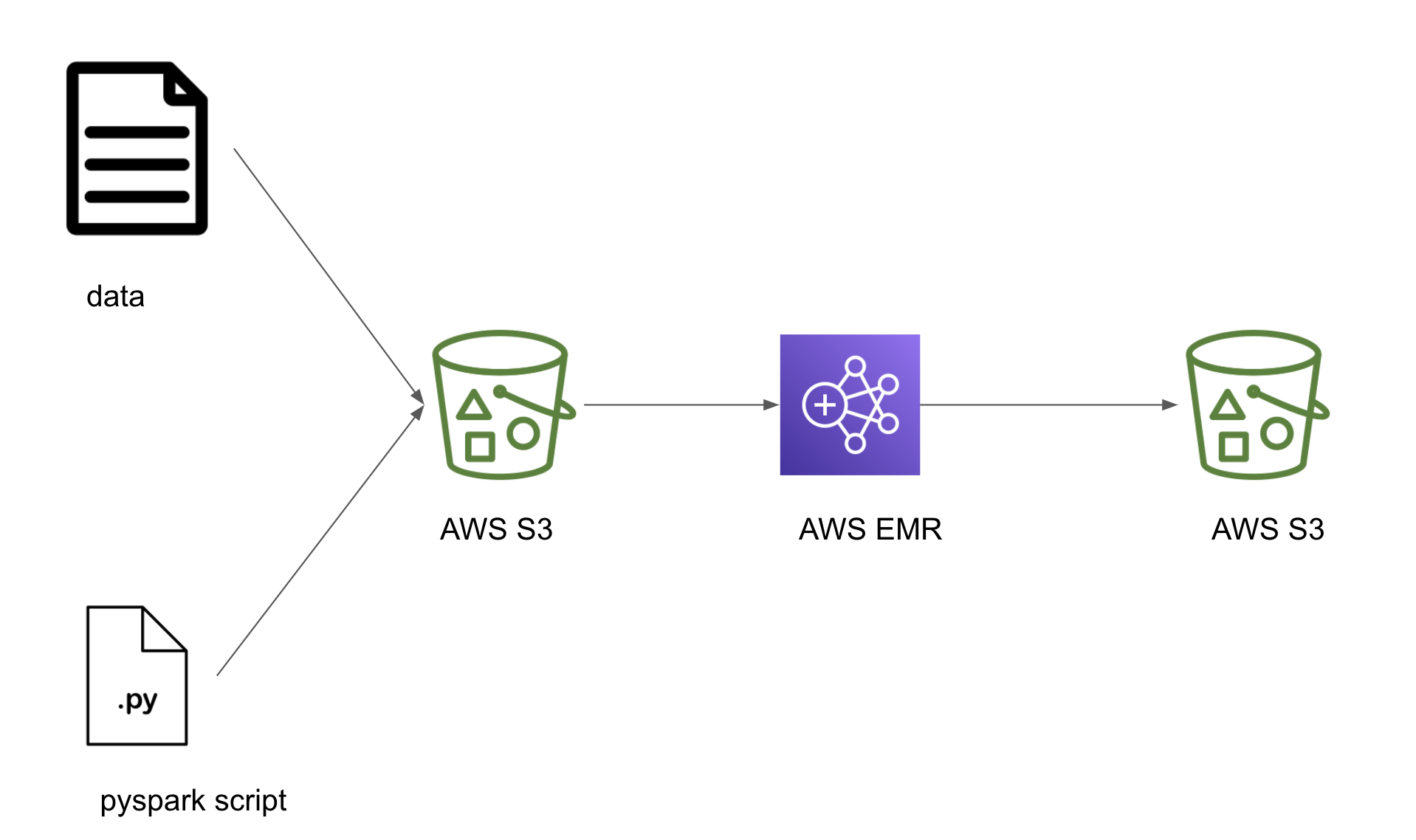
Design
Let’s build a simple DAG which uploads a local pyspark script and some data into a S3 bucket, starts an EMR cluster, submits a spark job that uses the uploaded script in the S3 bucket and when the job is complete terminates the EMR cluster. If you have an always-on spark cluster you can skip the tasks that start and terminate the EMR cluster.
Setup
Prerequisites
- docker (make sure to have docker-compose as well).
- git to clone the starter repo.
- AWS account to set up required cloud services.
- Install and configure AWS CLI on your machine.
If this is your first time using AWS, make sure to check for presence of the EMR_EC2_DefaultRole and EMR_DefaultRole default role as shown below.
aws iam list-roles | grep 'EMR_DefaultRole\|EMR_EC2_DefaultRole'
# "RoleName": "EMR_DefaultRole",
# "RoleName": "EMR_EC2_DefaultRole",
If the roles not present, create them using the following command
aws emr create-default-roles
Also create a bucket, using the following command.
aws s3api create-bucket --acl public-read-write --bucket <your-bucket-name>
Throughout this post replace <your-bucket-name> with your bucket name. eg.) if your bucket name is my-bucket then the above command becomes aws s3api create-bucket --acl public-read-write --bucket my-bucket. Press q to exit the prompt. In real projects the bucket would not be open to the public as shown at public-read-write.
Clone repository
Let’s start by cloning the repository and switching to the start-here branch.
git clone https://github.com/josephmachado/spark_submit_airflow.git
cd spark_submit_airflow
git checkout start-here
rm -r .git # if you want to remove the git reference and initialize your own
In this branch we will have DummyOperator for all the tasks, throughout this post they will be replaced with the actual operators required.
Get data
In your project directory, download the data and move it to the appropriate location as shown below.
mkdir ./dags/data
wget https://www.dropbox.com/sh/amdyc6z8744hrl5/AACZ6P5QnM5nbX4Gnk9_JW0Ma/movie_review/movie_review.csv?dl=0
mv movie_review* ./dags/data/movie_review.csv
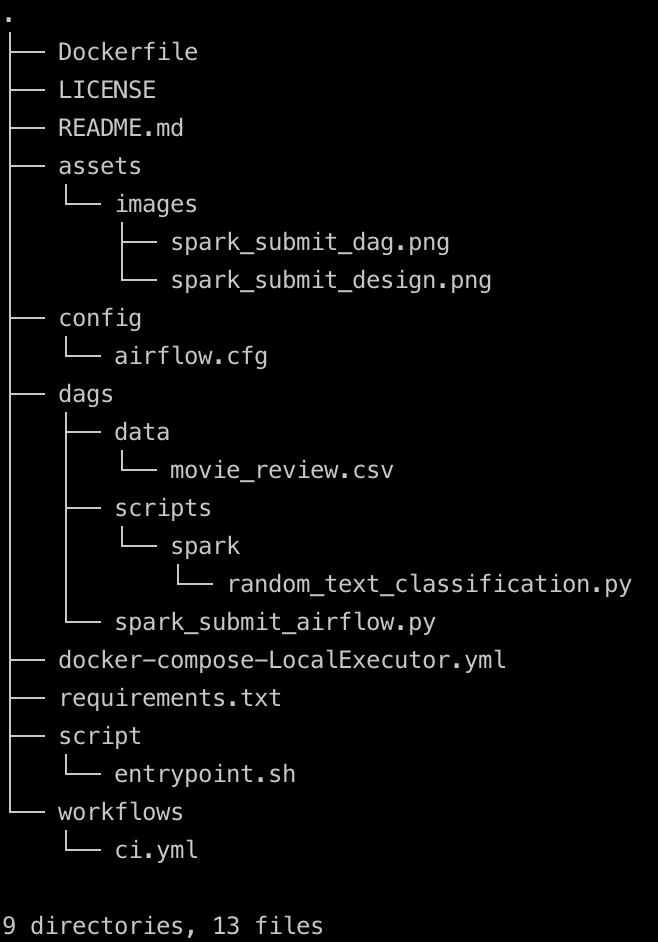
The project folder structure should look like shown below
Code
Let’s start implementing the following sections.
- Move data and script to AWS S3
- Create an EMR cluster
- Run jobs in the EMR cluster and wait for it to complete
- Terminate the EMR cluster
The random_text_classification.py is a naive pyspark script that reads in our data and if the review contains the word good it classifies it as positive else negative review. The code is self explanatory. The dag defined at spark_submit_airflow.py is the outline we will build on. This is a simple dag scheduled to run at 10:00 AM UTC everyday.
Move data and script to the cloud
In most cases the data that needs to be processed is present in the AWS S3. We can also store our pyspark script in AWS S3 and let our spark job know where it is located. We use Apache Airflow’s S3Hook to connect to our S3 bucket and move the data and script to the required location.
from airflow.hooks.S3_hook import S3Hook
from airflow.operators import PythonOperator
# Configurations
BUCKET_NAME = "<your-bucket-name>"
local_data = "./dags/data/movie_review.csv"
s3_data = "data/movie_review.csv"
local_script = "./dags/scripts/spark/random_text_classification.py"
s3_script = "scripts/random_text_classification.py"
# helper function
def _local_to_s3(filename, key, bucket_name=BUCKET_NAME):
s3 = S3Hook()
s3.load_file(filename=filename, bucket_name=bucket_name, replace=True, key=key)
data_to_s3 = PythonOperator(
dag=dag,
task_id="data_to_s3",
python_callable=_local_to_s3,
op_kwargs={"filename": local_data, "key": s3_data,},
)
script_to_s3 = PythonOperator(
dag=dag,
task_id="script_to_s3",
python_callable=_local_to_s3,
op_kwargs={"filename": local_script, "key": s3_script,},
)
From the above code snippet, we see how the local script file random_text_classification.py and data at movie_review.csv are moved to the S3 bucket that was created.
create an EMR cluster
Let’s create an EMR cluster. Apache Airflow has an EmrCreateJobFlowOperator operator to create an EMR cluster. We have to define the cluster configurations and the operator can use that to create the EMR cluster.
from airflow.contrib.operators.emr_create_job_flow_operator import (
EmrCreateJobFlowOperator,
)
JOB_FLOW_OVERRIDES = {
"Name": "Movie review classifier",
"ReleaseLabel": "emr-5.29.0",
"Applications": [{"Name": "Hadoop"}, {"Name": "Spark"}], # We want our EMR cluster to have HDFS and Spark
"Configurations": [
{
"Classification": "spark-env",
"Configurations": [
{
"Classification": "export",
"Properties": {"PYSPARK_PYTHON": "/usr/bin/python3"}, # by default EMR uses py2, change it to py3
}
],
}
],
"Instances": {
"InstanceGroups": [
{
"Name": "Master node",
"Market": "SPOT",
"InstanceRole": "MASTER",
"InstanceType": "m4.xlarge",
"InstanceCount": 1,
},
{
"Name": "Core - 2",
"Market": "SPOT", # Spot instances are a "use as available" instances
"InstanceRole": "CORE",
"InstanceType": "m4.xlarge",
"InstanceCount": 2,
},
],
"KeepJobFlowAliveWhenNoSteps": True,
"TerminationProtected": False, # this lets us programmatically terminate the cluster
},
"JobFlowRole": "EMR_EC2_DefaultRole",
"ServiceRole": "EMR_DefaultRole",
}
# Create an EMR cluster
create_emr_cluster = EmrCreateJobFlowOperator(
task_id="create_emr_cluster",
job_flow_overrides=JOB_FLOW_OVERRIDES,
aws_conn_id="aws_default",
emr_conn_id="emr_default",
dag=dag,
)
The EmrCreateJobFlowOperator creates a cluster and stores the EMR cluster id(unique identifier) in xcom, which is a key value store used to access variables across Airflow tasks.
add steps and wait to complete
Let’s add the individual steps that we need to run on the cluster.
from airflow.contrib.operators.emr_add_steps_operator import EmrAddStepsOperator
s3_clean = "clean_data/"
SPARK_STEPS = [ # Note the params values are supplied to the operator
{
"Name": "Move raw data from S3 to HDFS",
"ActionOnFailure": "CANCEL_AND_WAIT",
"HadoopJarStep": {
"Jar": "command-runner.jar",
"Args": [
"s3-dist-cp",
"--src=s3://{{ params.BUCKET_NAME }}/data",
"--dest=/movie",
],
},
},
{
"Name": "Classify movie reviews",
"ActionOnFailure": "CANCEL_AND_WAIT",
"HadoopJarStep": {
"Jar": "command-runner.jar",
"Args": [
"spark-submit",
"--deploy-mode",
"client",
"s3://{{ params.BUCKET_NAME }}/{{ params.s3_script }}",
],
},
},
{
"Name": "Move clean data from HDFS to S3",
"ActionOnFailure": "CANCEL_AND_WAIT",
"HadoopJarStep": {
"Jar": "command-runner.jar",
"Args": [
"s3-dist-cp",
"--src=/output",
"--dest=s3://{{ params.BUCKET_NAME }}/{{ params.s3_clean }}",
],
},
},
]
# Add your steps to the EMR cluster
step_adder = EmrAddStepsOperator(
task_id="add_steps",
job_flow_id="{{ task_instance.xcom_pull(task_ids='create_emr_cluster', key='return_value') }}",
aws_conn_id="aws_default",
steps=SPARK_STEPS,
params={ # these params are used to fill the paramterized values in SPARK_STEPS json
"BUCKET_NAME": BUCKET_NAME,
"s3_data": s3_data,
"s3_script": s3_script,
"s3_clean": s3_clean,
},
dag=dag,
)
last_step = len(SPARK_STEPS) - 1 # this value will let the sensor know the last step to watch
# wait for the steps to complete
step_checker = EmrStepSensor(
task_id="watch_step",
job_flow_id="{{ task_instance.xcom_pull('create_emr_cluster', key='return_value') }}",
step_id="{{ task_instance.xcom_pull(task_ids='add_steps', key='return_value')["
+ str(last_step)
+ "] }}",
aws_conn_id="aws_default",
dag=dag,
)
In the above code we can see that we specify 3 steps in the SPARK_STEPS json, they are
- copy data from AWS S3 into the clusters HDFS location
/movie. - Run a naive text classification spark script
random_text_classification.pywhich reads input from/movieand write output to/output. - Copy the data from cluster HDFS location
/outputto AWS S3clean_datalocation, denoted by the s3_clean configuration variable.
We get the EMR cluster id from xcom as shown in job_flow_id="{{ task_instance.xcom_pull(task_ids='create_emr_cluster', key='return_value') }}". We also get the last step id from xcom in our EmrStepSensor. The step sensor will periodically check if that last step is completed or skipped or terminated.
terminate EMR cluster
After the step sensor senses the completion of the last step, we can terminate our EMR cluster.
from airflow.contrib.operators.emr_terminate_job_flow_operator import (
EmrTerminateJobFlowOperator,
)
# Terminate the EMR cluster
terminate_emr_cluster = EmrTerminateJobFlowOperator(
task_id="terminate_emr_cluster",
job_flow_id="{{ task_instance.xcom_pull(task_ids='create_emr_cluster', key='return_value') }}",
aws_conn_id="aws_default",
dag=dag,
)
In the above snippet, we get the cluster id from xcom and then terminate the cluster.
Run the DAG
Let’s create our Apache Airflow instance as shown below from our project directory
docker-compose -f docker-compose-LocalExecutor.yml up -d
This will create the services needed to run Apache Airflow locally. Wait for a couple of minutes(~1-2min) and then you can go to http://localhost:8080/admin/
to turn on the spark_submit_airflow DAG which is set to run at 10:00 AM UTC everyday. The DAG takes a while to complete since
- The data needs to be copied to S3.
- An EMR cluster needs to be started(this take around 8 - 10min generally).
You can see the status of the DAG at http://localhost:8080/admin/airflow/graph?dag_id=spark_submit_airflow
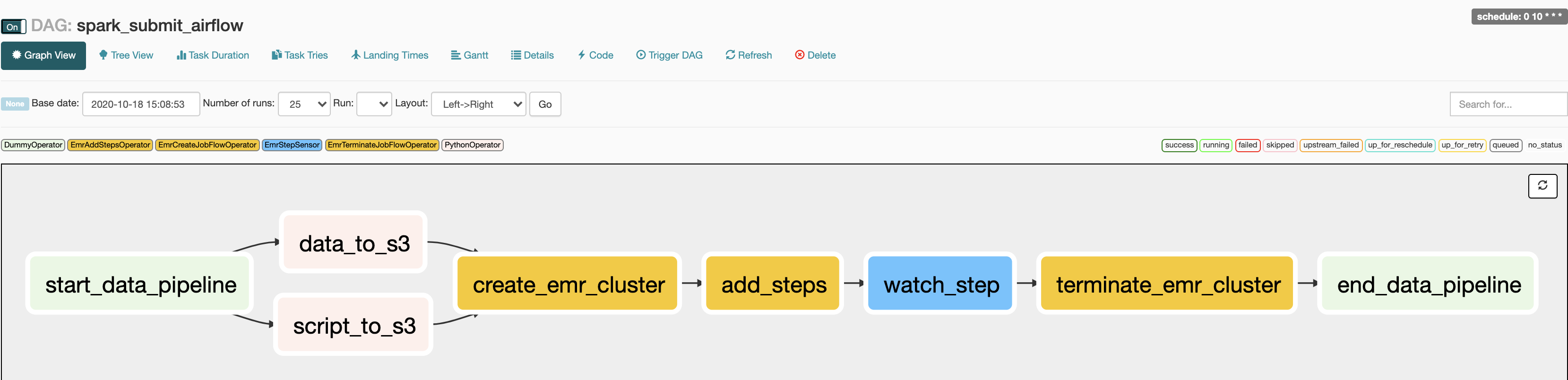
NOTE: If your job fails or you stop your Airflow instance make sure to check your AWS EMR UI console to terminate any running EMR cluster. You can also remove the S3 you created as shown below
aws s3 rm s3://<your-bucket-name> --recursive
aws s3api delete-bucket --bucket <your-bucket-name>
Press q to exit the prompt. You can spin down the local Airflow instance as shown below.
docker-compose -f docker-compose-LocalExecutor.yml down
Conclusion
Hope this gives you an idea of how to create a temporary AWS EMR clusters to run spark jobs. One of the biggest issues with this approach is the time it takes to create the EMR cluster. If you are using an always-on EMR cluster you can skip the tasks to create and terminate the EMR cluster. In the above code snippet the jobs will run serially, there may be cases where parallel runs of jobs may be better. In such cases you can use this , to schedule steps in parallel. If you have any questions or comments please leave them in the comment section below.